Abstract
Introduction: Multiple Myeloma (MM) is a plasma cell malignancy characterized by high polyclonality, heterogeneity of treatment response, and long-term adaptation to treatment. The apparent complexity of underlying biological networks of MM necessitates a systems approach to its study. Advances in instrumentation, machine learning, and computing make the systems approach possible and enable novel insights into MM disease drivers, patient subpopulations, and potential therapies.
We have developed a high-dimensional network model of MM based on data from 645 patients (pts) in the Interim Analysis 9 (IA9) MMRF CoMMpass trial dataset (NCT0145429). This model, developed using the REFS™ causal inference engine, consists of an ensemble of 256 Bayesian networks, each representing the inferred causal relationships between 30,084 clinical and genomic variables. The results from this model include the identification of a pathway driving high-risk disease (progression or death within 18 months) and the characterization of a subpopulation of patients with increased progression-free survival (PFS) after stem cell transplantation (SCT). We have now tested the overall IA9 model and its key results using the IFM/DFCI 2009 dataset (NCT01191060), as well as performed experimental pre-clinical validation of core molecular drivers in the high-risk status pathway.
Methods: RNA-Seq, demographic and clinical data modalities in the IFM/DFCI dataset were processed and normalized using the same pipeline as the IA9 dataset. Among the 30,084 variables in the IA9 model, 24,559 were present in the IFM/DFCI dataset, and a total of 323 pts had complete clinical and molecular data.
Results: The IA9 model contains 121,708 edges (causally enriched statistical associations) that appear in at least 25% of the inferred networks in the ensemble. 93,636 of these edges (77%) were tested in the IFM/DFCI dataset and 81,155 edges (87%) had significant q-value (< 0.05) and effect sizes of the same sign between the two datasets. The effect sizes between datasets were highly correlated: among all edges, Pearson's r = 0.89; among validated edges, r = 0.93.
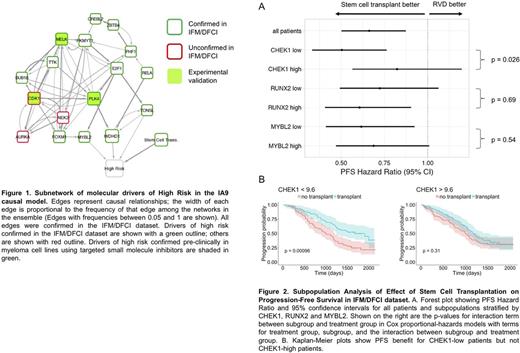
With regards to predictors of high risk disease, the IA9 model contains a subnetwork of 17 RNA-Seq variables driving high-risk status, notably involving cell-cycle regulators CDK1, MELK and PLK4, known targets of inhibitor drugs (Figure 1). In order to verify the robustness of these potential drivers, we tested the association between their gene expression and high-risk status in the IFM/DFCI dataset, including SCT as a covariate. We performed logistic regression for each of the variables, and 13 genes, including MELK, FOXM1, E2F1 and PLK4, maintained statistical significance (q < 0.05). The functional relevance of these potential drivers of high risk was confirmed pre-clinically in myeloma cell lines using targeted small molecule inhibitors of MELK, CDK1 and PLK4.
Simulation of the IA9 model also revealed a patient subpopulation with increased PFS in response to SCT and decreased PFS in its absence. The top driver of this subpopulation was expression of CHEK1; other drivers included RUNX2 and MYBL2. We examined these genes in the IFM/DFCI dataset by defining cohorts of pts with high or low gene expression (above or below median). For the full cohort, the PFS Cox Hazard Ratio (HR) was 0.66, with 95% Confidence Interval (CI) 0.50-0.87. For CHEK1-low pts, a large PFS benefit was observed in response to SCT (HR=0.51; 95% CI 0.33-0.76, p=0.001), whereas no benefit was observed for CHEK1-high pts (HR=0.82; 95% CI 0.57-1.20, p=0.31). Cox survival modeling of PFS revealed a statistically significant interaction between CHEK1 expression and SCT (p = 0.026). RUNX2 and MYBL2 subpopulations showed only modest differences in HR (Figure 2).
Conclusions: Together, these results confirm key predictive results of the IA9 computational model in an out-of-sample dataset. This model should now help researchers to focus on the most promising targets and pathways, as well as to address unanswered questions and unmet needs in myeloma, especially high risk disease.
Furchtgott: GNS Healthcare: Employment. Gruber: GNS Healthcare: Employment. Attal: Sanofi: Consultancy; JANSSEN: Consultancy, Research Funding; Amgen: Consultancy, Research Funding; Celgene: Consultancy, Research Funding. Moreau: Celgene, Janssen, Takeda, Novartis, Amgen, Roche: Membership on an entity's Board of Directors or advisory committees; Celgene: Consultancy, Honoraria; Takeda: Honoraria; Novartis: Consultancy, Honoraria; Amgen: Honoraria; Bristol-Myers Squibb: Honoraria; Janssen: Consultancy, Honoraria; Millennium: Consultancy, Honoraria; Onyx Pharmaceutical: Consultancy, Honoraria. Avet-Loiseau: Celgene, Janssen, Amgen, Bristol-Myers Squibb, Sanofi: Honoraria, Speakers Bureau; Janssen, Sanofi, Celgene, Amgen: Consultancy; Celgene, Janssen: Research Funding. Runge: GNS Healthcare: Employment. Wuest: GNS Healthcare: Employment. Rich: GNS Healthcare: Employment. Khalil: GNS Healthcare: Employment. Hayete: GNS Healthcare: Employment. Ludwig: AMGEN: Consultancy, Research Funding, Speakers Bureau; Takeda: Research Funding, Speakers Bureau; Takeda: Consultancy, Research Funding, Speakers Bureau; Celgene: Speakers Bureau; Bristol-Meyers: Speakers Bureau; Janssen-Cilag: Consultancy, Speakers Bureau.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal